
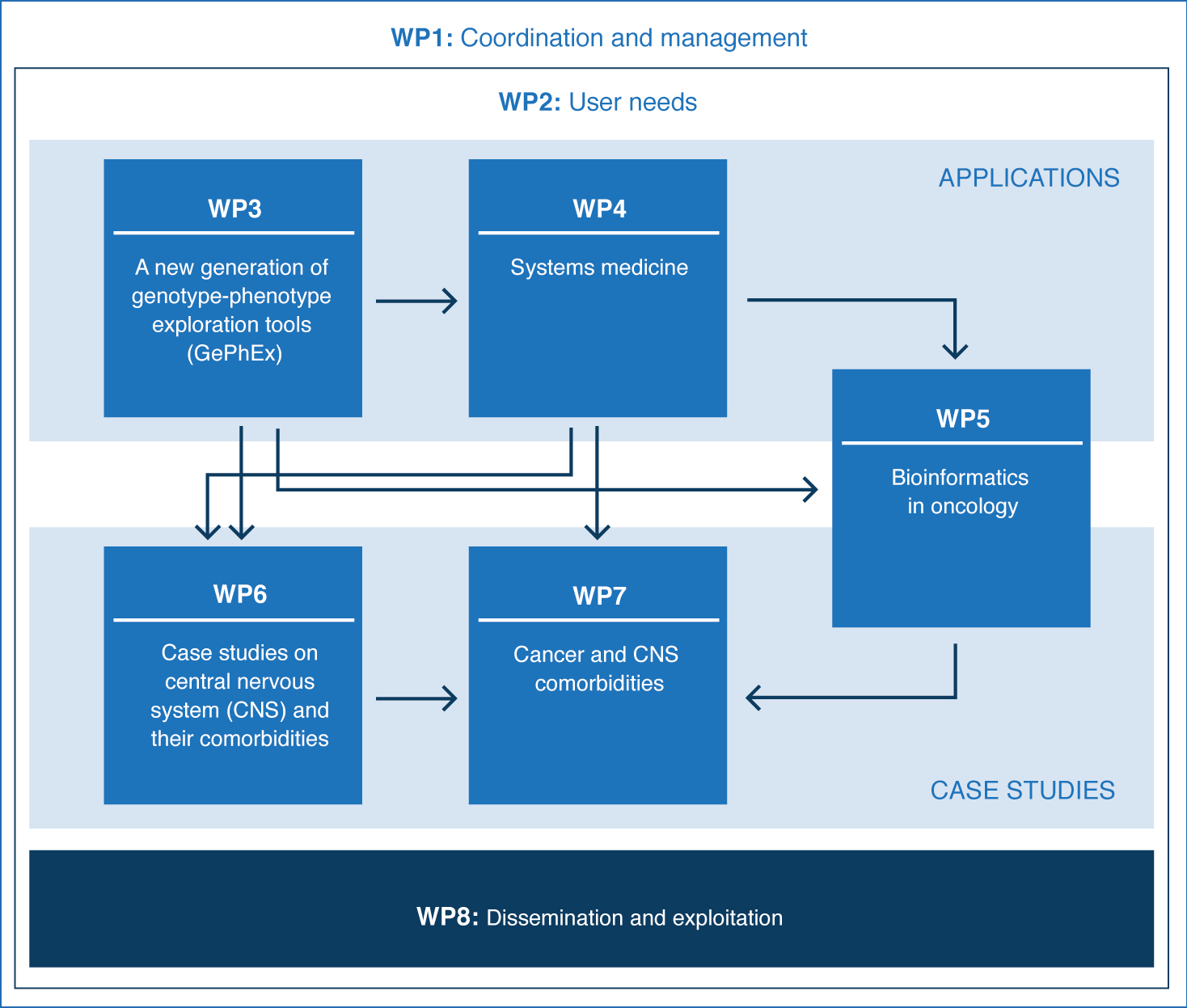
PROJECT STRUCTURE
The work plan structure in MedBioinformatics has been carefully designed to cover all aspects requiring specific effort towards a successful completion, and divides activities into eight work packages:
START: Month 1
END: Month 36
WORK PACKAGE LEADER: PSMAR
END: Month 36
WORK PACKAGE LEADER: PSMAR
Objectives:
- To provide the overall scientific direction and to drive the progress of the project, steering efforts of the partners for the achievement of the project’s objectives and milestones, and ensuring that the work is undertaken with appropriate quality levels.
- To set-up a project management structure that ensures an efficient operational management, including administrative, financial and legal issues, as well as appropriate liaison with the European Commission.
- To ensure that the project is appropriately managed according to the work plan, supporting the Coordinator in organizing and supervising the work.
- To provide resources, procedures and tools for ensuring that all results are delivered on time, with an adequate quality level and within cost, including quality control procedures on deliverables and interim and final review of the project achievements from a management perspective.
- To enable the appropriate communication and work dynamics to help drive the whole Consortium as a team towards successful completion.
START: Month 1
END: Month 36
WORK PACKAGE LEADER: BMD
END: Month 36
WORK PACKAGE LEADER: BMD
Objectives:
- To create and promote software development guidelines in translational bioinformatics.
- To gather interactively and analyse the user requirements to leverage effective biomedical applications.
- To assess the software tools developed in the framework of the project.
START: Month 1
END: Month 36
WORK PACKAGE LEADER: UPF
END: Month 36
WORK PACKAGE LEADER: UPF
Objectives:
- To implement, develop and benchmark new algorithms linking genotypes and phenotypes that go beyond the independent analysis of single variants.
- To implement, develop and benchmark algorithms predicting individual disease risks and treatment outcomes based on genome and biomarker data.
- To ease the interpretation of genotype-phenotype links by improving and integrating databases on known genetic interactions, genotype-phenotype associations and the functional effect of genomic variants.
- To integrate all the developments above into GePhEx, a desktop-able application with a steep learning curve that can be used by clinicians and clinical researchers to link genomes and phenomes at different levels.
START: Month 1
END: Month 36
WORK PACKAGE LEADER: UCPH
END: Month 36
WORK PACKAGE LEADER: UCPH
Objectives:
- To develop a Disease Biomarker Browser (DBB) that gathers information about disease biomarkers from public sources and will display it in a way that will allow the user to easily assess the translational potential of these biomarkers.
- To develop a Disease Trajectory Comorbidity Browser (DTCB) that provides a wide-angle, comprehensive view of disease comorbidity from different perspectives (molecular, phenotypic and population levels). It will provide quantitative risk measures for comorbidities based on disease prevalence data, as well as their plausibility given certain molecular features (based on current knowledge of disease mechanisms) and phenotypic information. The DTCB will also make use of information on the temporal relationships among the clinical events.
START: Month 1
END: Month 36
WORK PACKAGE LEADER: UPF
END: Month 36
WORK PACKAGE LEADER: UPF
Objectives:
- To develop software tools that help cancer researchers and clinical oncologists interpreting the mutations of a tumour genome.
- To develop computational methods and tools for predicting the most effective drug combination for a given cancer patient.
START: Month 3
END: Month 36
WORK PACKAGE LEADER: PSMAR
END: Month 36
WORK PACKAGE LEADER: PSMAR
Objectives:
- To apply the MedBioinformatics methods and tools to:
- Identification of differential biomarkers between Major Depression and Alcohol-induced Major Depression.
- Gathering and prioritization of biomarkers for Alzheimer Disease from the scientific literature and GWAS studies.
- Study of the disease trajectories for the comorbidity between Alzheimer Disease and depression and generation of plausible biological hypothesis to explain this comorbidity.
START: Month 10
END: Month 34
WORK PACKAGE LEADER: PSMAR
END: Month 34
WORK PACKAGE LEADER: PSMAR
Objectives:
- To apply the MedBioinformatics methods and tools to:
- Study of the inverse comorbidity between Alzheimer Disease (AD) and Cancer.
- Study of the comorbidity between Major Depression (MD) and Cancer.
- Study of the effect of chronic pharmacologic treatment in cancer-CNS comorbidities.
START: Month 1
END: Month 36
WORK PACKAGE LEADER: SYNAPSE
END: Month 36
WORK PACKAGE LEADER: SYNAPSE
Objectives:
- To design a plan that allows for optimal communication within the project Consortium and the dissemination of information and knowledge generated by the project to the scientific community, other relevant stakeholders and the society at large.
- To develop a communication plan for efficiently achieving the aforementioned dissemination objectives.
- To design and produce the communication tools that will be needed to implement the communication plan.
- To analyse the context of use of the tools developed in the context of MedBioinformatics in order to gather critical information for the business plan.
- To devise sustainability models for the MedBioinformatics derived from the analysis above, which ensures the sustained exploitation of the methods and tools developed during the project.
